Whole Slide Images (WSI)
Large histopathlogy images such as H & E stains can be visualized in rakaia v0.23.0 and later alongside multiplxed image blends from both antibody-based assays and spatial assays. depending on the source of the multiuplexed blend in the current session, rakaia may also be able to align the WSI coordinates to the current blend (currently this applies only to 10X assays such as Visium and Xenium).
WSI rendering requirements
Viewing WSIs in rakaia requires an installation of libvips; without a standlone vips installation, rakaia will inform the user that WSIs cannot be processed and rendered with openseadragon. Users should fetch the relevent vips installation for their OS from the installation page, as this library is not shipped/installed in rakaia by default.
WSI viewer tab
WSIs are viewed in a separate tab from the multiplexed blend; this can be toggled by switching between the Canvas and WSI sub tabs in the main canvas page:
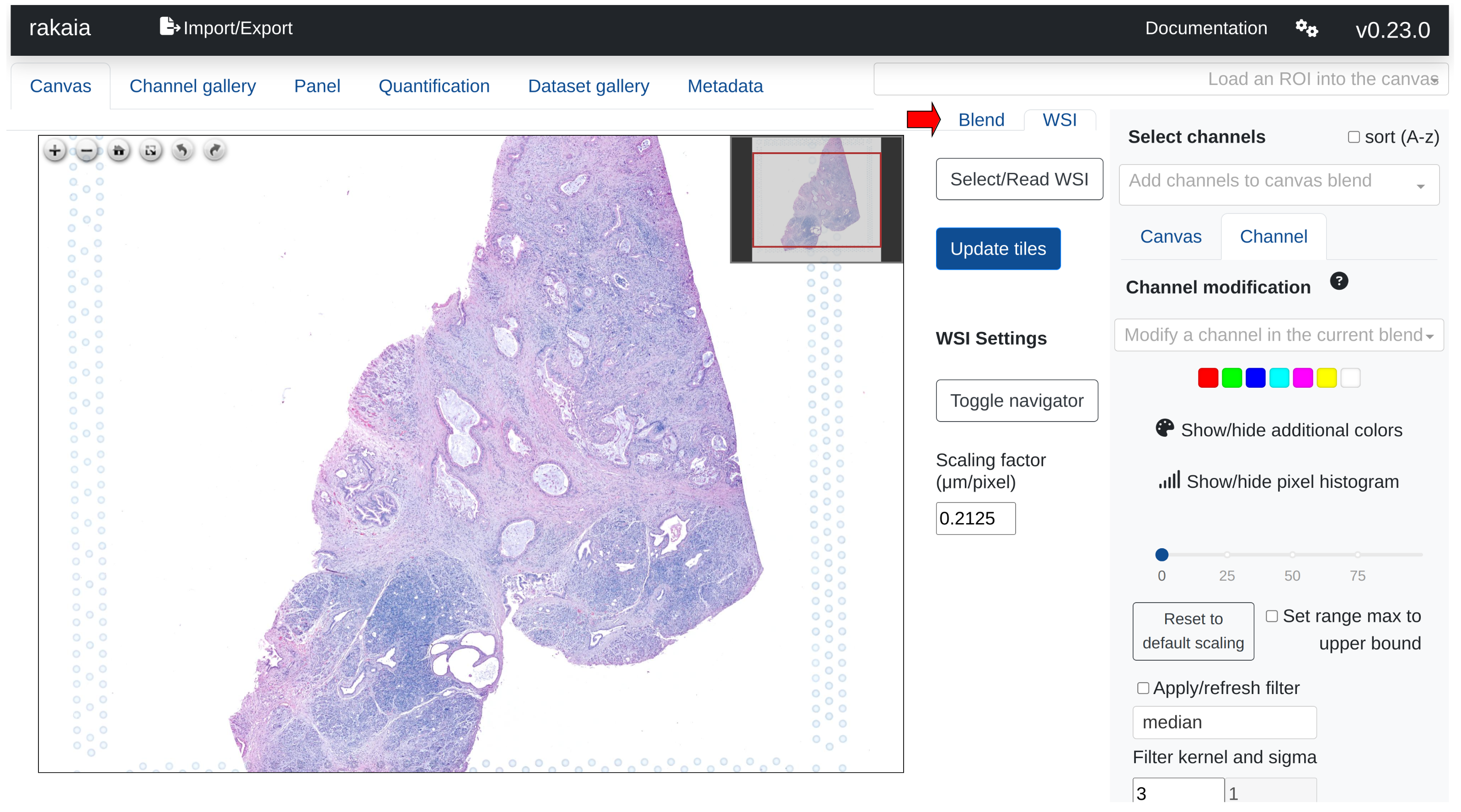
Users can import WSIs using either a direct file path import under Select/Read WSI from the tab as shown above, or through drag and drop under the Import/Export side tab, Show/hide additional imports -> Choose or drop WSI.
rakaia currently supports the following image formats that are converted in dzi tiles for rendering:
- tiff (pyramidal)
- svs
- btf (big TIFF)
Users can switch the WSI loaded into the WSI viewer using the Select/Read WSI -> Select WSI dropdown. Once a selection is made, a loader will start indicating that the dzi tiles for the selected WSI are being generated. Once finished, users should select Update tiles to finish rendering the WSI in the viewer.
Users must select Update tiles after every new WSI selection, as rakaia oes not explicitly listen for the newly generated tiles.
openseadragon viewer
Viewing WSIs is supported by openseadragon, a Javascript library designed to render high-resolution, zoomable images. The viewwer enables zooming, panning, and full-screen views of WSIs. Visit the osd documentation for more information.
Aligning WSIs to spatial datasets
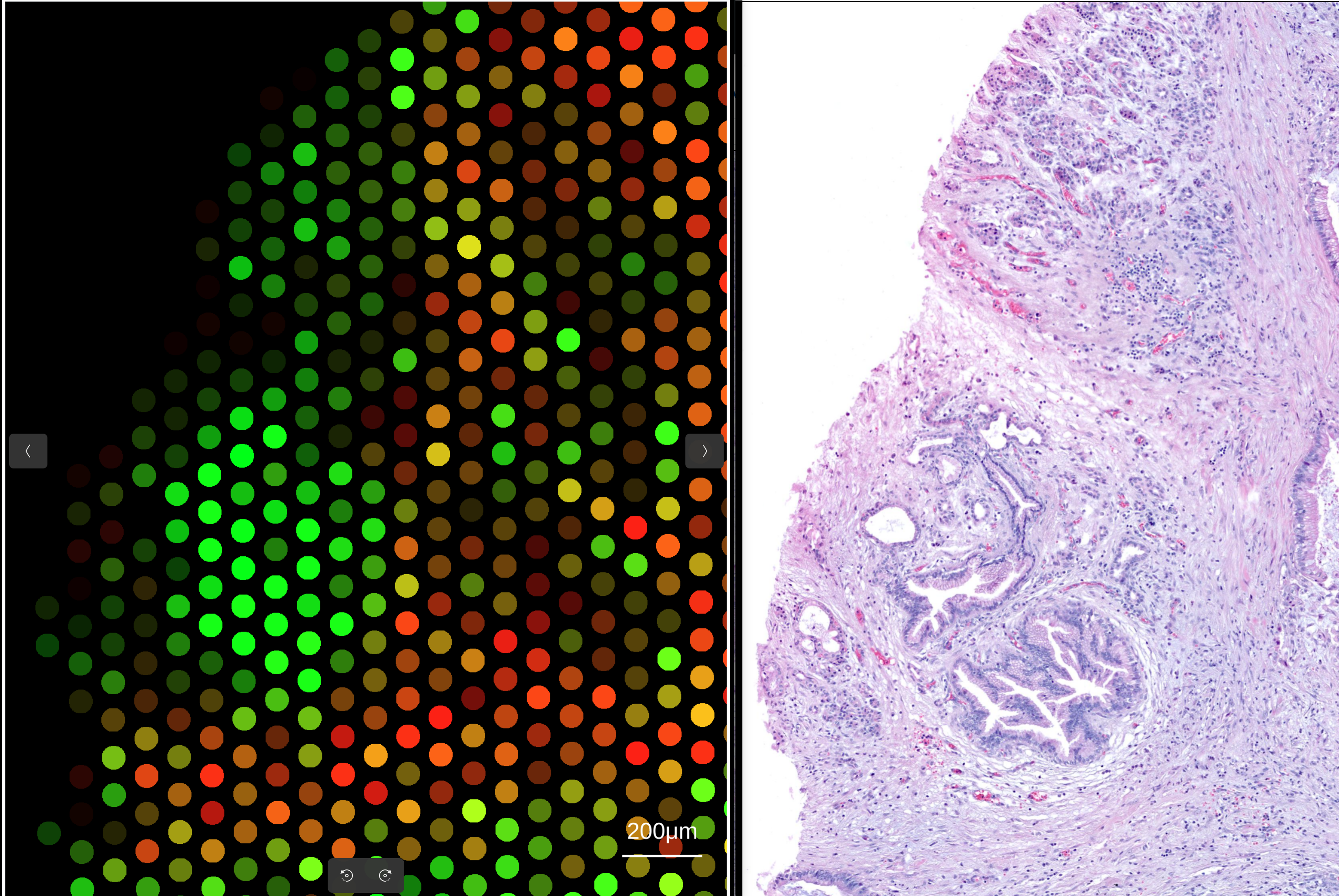
rakaia curently supports WSI alignment to both 10X Visium and 10X Xenium datasets. 10X Visium alignment to WSIs is straightforward and requires no additional data processing or file import, as the assay by default retains the global coordinates from the WSI when spots are rendered.
To align the 10X Visium to its matched WSI, simply import a 10X Visium assay into the main canvas tab (see here for importing Visium data) and the corresponding WSI into the osd viewer. Once a Visium blend is made, simply zooming in on a subset of the spots in the main canvas will automatically transfer the coordinate bounds into the WSI view:
Aligning 10X Xenium with a transformation matrix
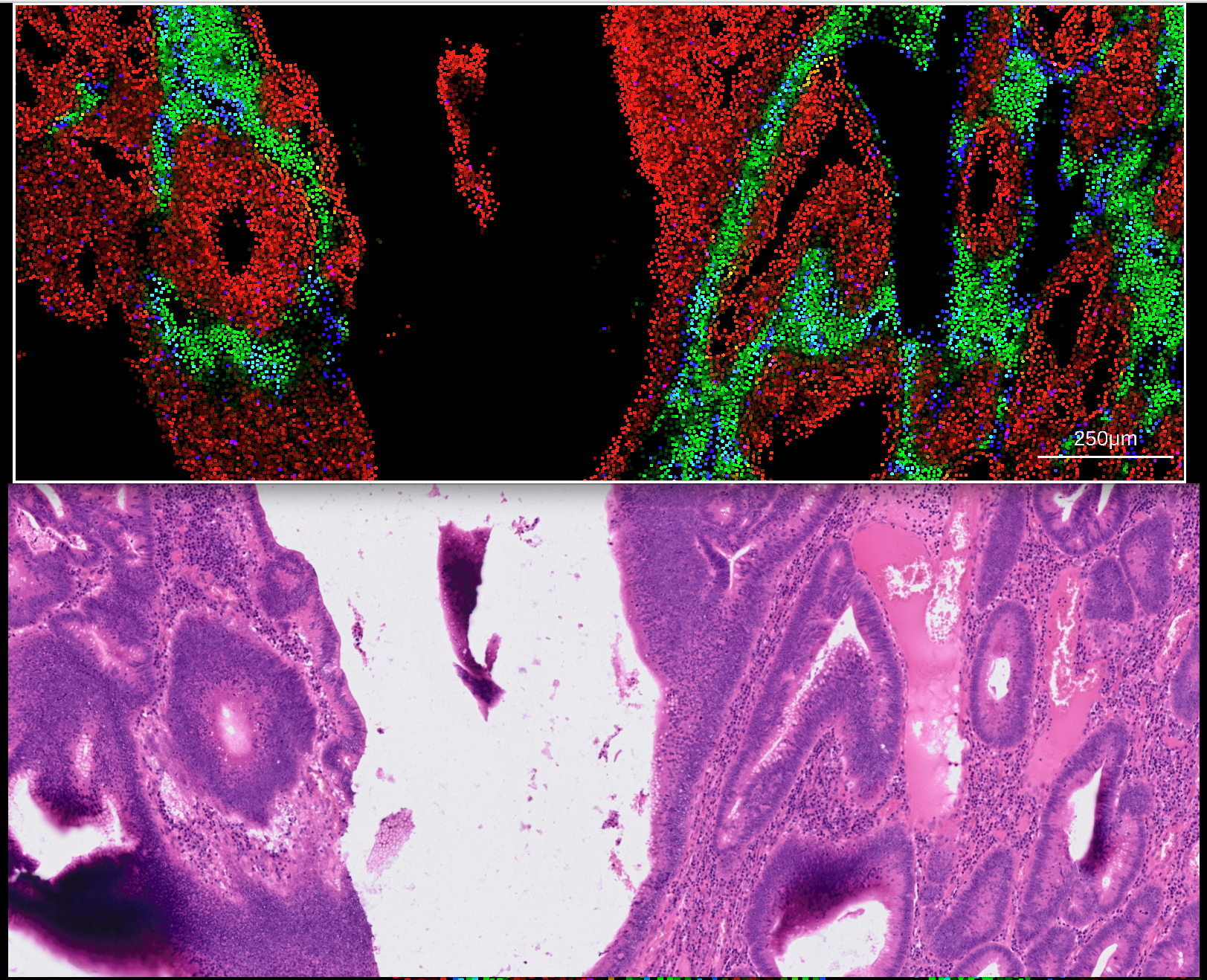
Aligning 10X Xenium datasets requires an additional import step of an affine transformation matrix generated from Xenium explorer. Visit this link for 10X vendor information on how to generate a 3x3 affine transformation matrix.
The transformatiom matrix should be exported in CSV format, and can then be imported into rakaia under File import -> Show/hide additional imports -> Choose or drop affine transformation matrix for WSI. Similar to 10X Visium, both the expression h5ad and WSI for the Xeium assay should be imported into the session. With the addition of the transformation matrix, zooming in on a Xenium anvas blend will trigger an update in the matched WSI:
Xenium image scaling factors
When aligning 10X Xenium WSIs, it is important that the user know which image level/series the WSI is derived from, and set the scaling factor accordingly in the WSI tab. By default, the WSI is assumed to be from the top level (series 0), which has an image factor of 0.2125 (this is the default rakaia value). To change the Xenium scale factor, visit the link below: