Sessions, Panel Metadata, & Naming
rakaia sessions are defined as browser instances of the application where the data imports are reset to blank templates, allowing any compatible
datasets to be read in. A page refresh qualifies as as new rakaia session, as the Serverside objects containing imaging and supporting data are reset when this occurs.
Data imports into rakaia are accompanied by the generation of parsed or auto-generated metadata variables that describe the parameters of the datasets being analyzed. Below is a summary of the different metadata information available to the user and what parameters may be edited in-session.
Session compatibility
rakaia sessions are defined by datasets that share a common biomarker panel; that is, images and quantification objects that have measured common entities. it is important to note that currently, rakaia does not support sessions for different biomarker panels. There are several reasons for this design decision, including an increased possibility of session errors and challenges with establishing gene aliases matches between channel names and their in-session labels. Therefore, users will need to ensure that imported datasets share a common panel, or they may face a PanelLengthMismatch error (See Troubleshooting for more information) and the session will not proceed.
When data are imported, the first file imported will be parsed for the biomarker panel. This file will set the internal names for the channel names (keys used by rakaia to retrieve images from data files) as well as labels (the way that the biomarker appears in the rakaia session components and canvas). When parsing from mcd, the names will be set from the channel_names attribute for each acquisition, and labels are set similarly from channel_labels. Parsing files such as tiff stacks, which often do not provide schematic metadata on the channels, will create a generic template where each biomarker/channel is named and labelled with the same generic channel name, channel_#', where the number is the index of the channel in the stack. The labels can be edited within the session (See editable metadata).
By default, rakaia will import the list of filenames by natural sorting (A-z); this can be disabled by the user so that the first file imported will be generated from the selection in the specific widget/component.
Data information preview
The dataset information preview table provides a summary of the each dataset ROI in the current session, with a naming convention that includes the filename/experiment, slide number (if applicable to the input filetype), and a name identifier for the particular ROI:
Additional columns include the ROI dimensions and the length of the panel (number of channels) associated with each ROI. Specific ROIs are selectble from the leftmost input column of the preview table.
Internal naming conventions
By default, rakaia will create a string representation of specific ROIs using a combination of the experiment/filename, slide number,
and acquisition id, if provided. Specific file formats such as mcd will have a standard naming convention of filename+++slide_number+++acquisition_name, where the filename and acquisition name are created by the user on the IMC machine. MCD import assumes that there can
be multiple experiments (filenames) in a session, and within each file, multiple slides coresponding to multiple ROIs. When an mcd file is parsed
into a session, all of the slides are ROIs are imported.
Tiff import is adjusted slightly as compared to mcd import as there is no guarantee of experiment and acquisition names being contained in
any tiff-associated metadata. rakaia makes no distinction between ome.tiff and standard tiff formats, as the convention for metadata is very broad for the former format. Therefore, tiff import follows these conventions:
- filenames provide the experiment name
- each tiff file should correspond to one ROI. If the ROI has multiple channels, each channel should be a page in the tiff
- slide number is always 0 (just acts as a placeholder)
- each acquisition in the tiff collection is named consistently as 'acq', so an import of three filenames could look like
filename1+++slide0+++acq,filename2+++slide0+++acqandfilename3+++slide0+++acq. Because rakaia uses the basename filename as the experiment identifier, the imported ROI identifiers will remain unique as long as the filenames are unique.
String delimiter
rakaia autogenerates internal string identifiers for each ROI that sequentially hold the experiment/base filename, slide number, and acquisition
identifier. These elements are separated by a special delimiter, which by default is +++; rakaia will perform string splitting at various steps in
the workflow to pull the necessary information from an internal ROI identifier, such as the base filename or the acquisition name.
The default delimiter of +++ can be changed if the user has the + character in any portion of the filename or acquisition id. For example, in an mcd file where one of the ROIs has been named +(Positive_control), the string representation of the dataset would be filename+++slide0++++(Positive_control), causing a string split error when using the default +++. Another recommended string delimiter that preserves readability of the dataset is ---. When used above, the string splitting property is now preserved: filename---slide0---+(Positive_control). The string delimiter may be set under Set additional session variables next to the documentation link in the header.
Editable panel metadata
rakaia provides an editable metadata table under Panel tab where the names and labels of the biomarker panel for the current session can either be re-configured, edited, or downloaded. When the metadata panel is parsed, rakaia will produce a copy of the Channel label column named rakaia Label, which is editbale for the particular biomarker. Editing the cells in this colun will produce a new name for the channels that will propogate to the internal dropdown menus as well as the channel label in the canvas legend. This is referred to as the marker alias within the session.
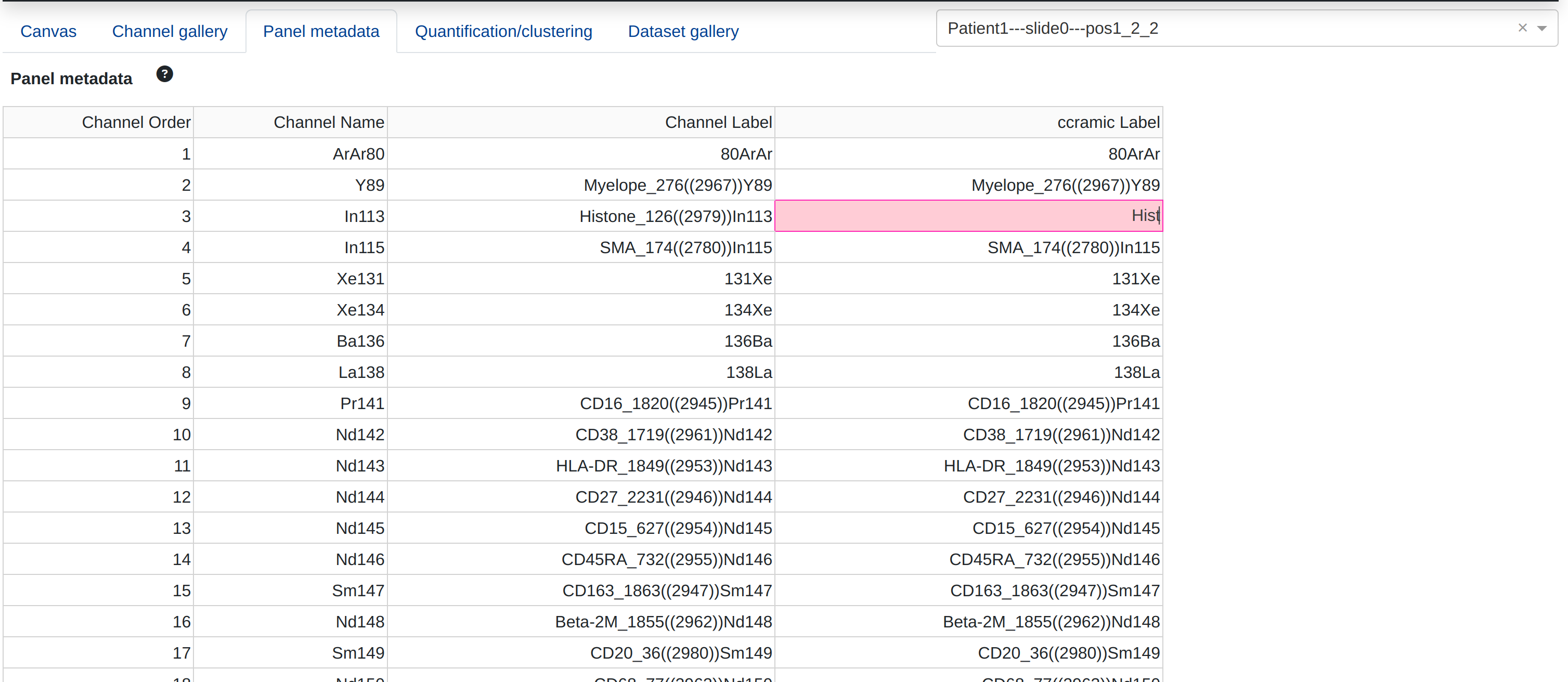
In the configuration above, the channel that is internally identified In113 will be displayed visually as Hist, corredponding to a histone marker. Label changes can be reverted at any point in the session, and labels can also be bulk-edited by copying the entire column of labels from a CSV template into the rakaia Label column by double clicking the first column and pasting. Note that the user must not copy the header for this to work, as the number of elements must align properly to provide a gene alias to each biomarker.