Models & Plugins
rakaia supports a variety of classification models and algorithms for labelling and stratifying elements of imaging datasets. Currently, models that process quantified object expression results are supported, with plans for pixel-level classifiers to be supported in the future.
Object-level models
Current object-level classification models in rakaia can be found under the Quantification tab with the Plugins button. The algorithms include:
- Leiden clustering
- Random forest classification for pre-annotated object/structures
- Object mixing/segmentation error classifier
Information on each model can be found below
Leiden clustering
Leiden clustering is a common community detection algorithm used to group similar objects together. rakaia allows leiden clustering to identify key object communities that may have shared biological function or importance, based on the marker expression from quantification results. Visit the link here for more information on the leiden algorithm.
Random forest classification
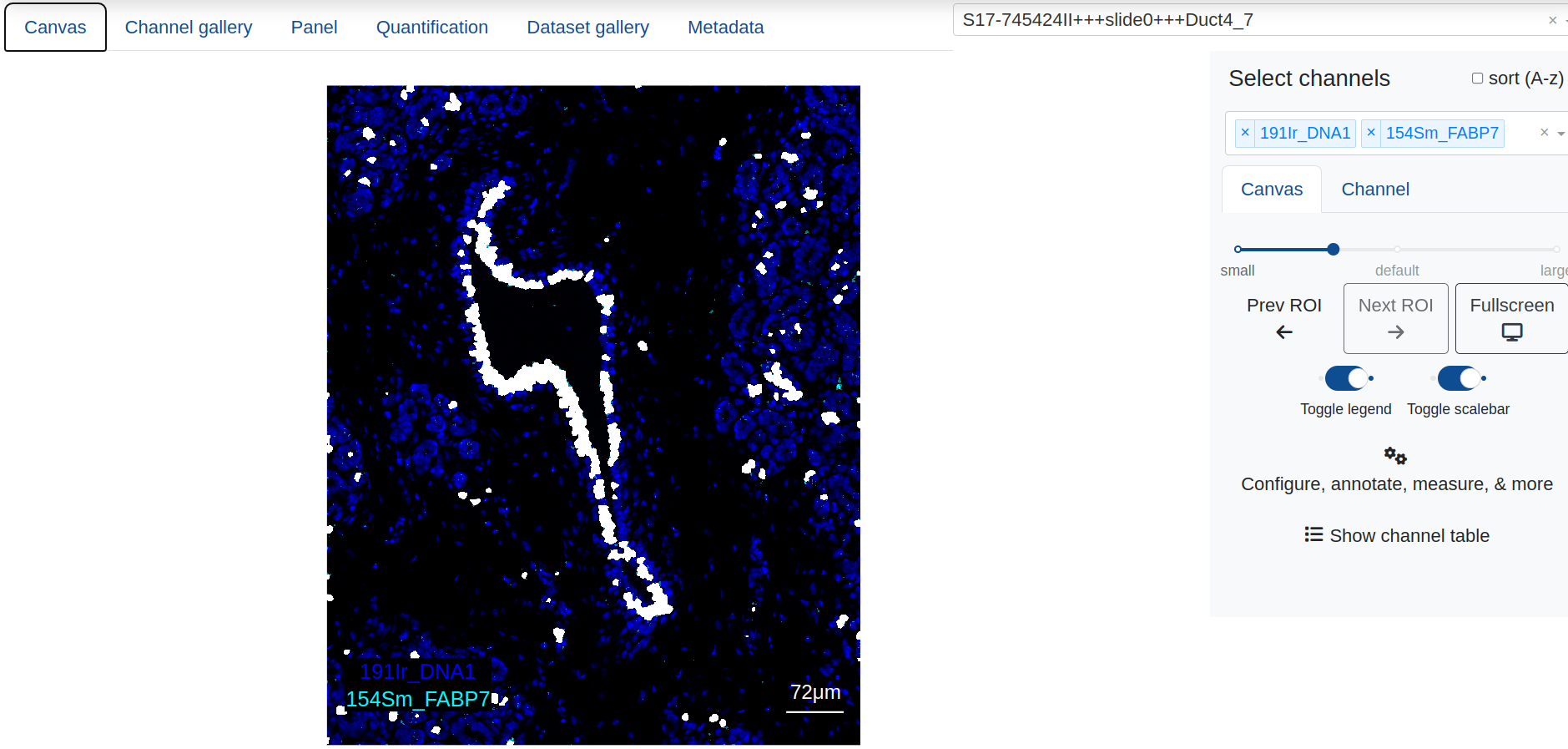
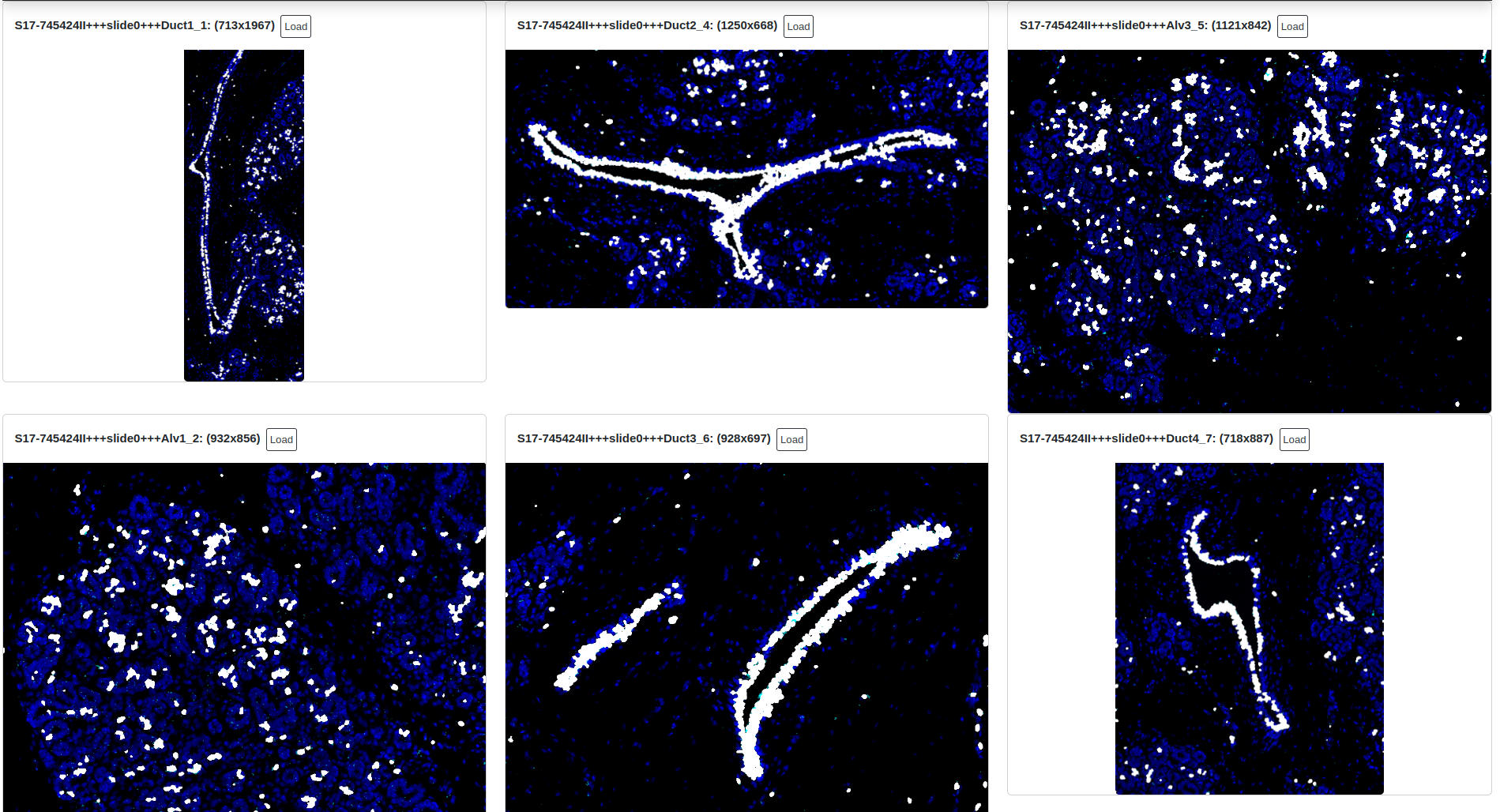
Users may choose to annotate the objects and structures in a specific ROI and then search for similar objects across all quantified ROIs in the session. This can be achieved using the random forest classifier, which will use any ROIs that have existing annotation labels for training and then classify the entire data set.
In an example below, an interesting biological structure is identified using manual object gating (visit the section on quantifying and gating for information on how to do this). Once an annotation is made, the annotation is supplied to the classifier, and the random forest then classifies every object across all other ROIs (6) as either being part of a similar region, or unassigned
Object mixing
The object mixing classifier performs similarly to the random forest model above, but will identify objects that are likely to be mis-segmented or have expression statistics that indicate mixing among teo or more objects. The model will generate synthetically mixed cells from a random subset of the provided quantificatoin results, creating a balanced training set that contains both true and known mixed cells. The classifier will then output either 0 (not mixed) or 1 (mixed) labels for every object, indicating the its most likely label based on its overall marker expression. Identifying potentially poorly segmented objects or those that may have expresison mixtures could prove to be a key step in postprocessing and quality control.
Model requirements
All models in rakaia require quantification results for one of more ROIs in the session. Visit the Measuring & Detecting section for information on importing quantification results from external pipelines, or for generating expression results per ROI in the application.
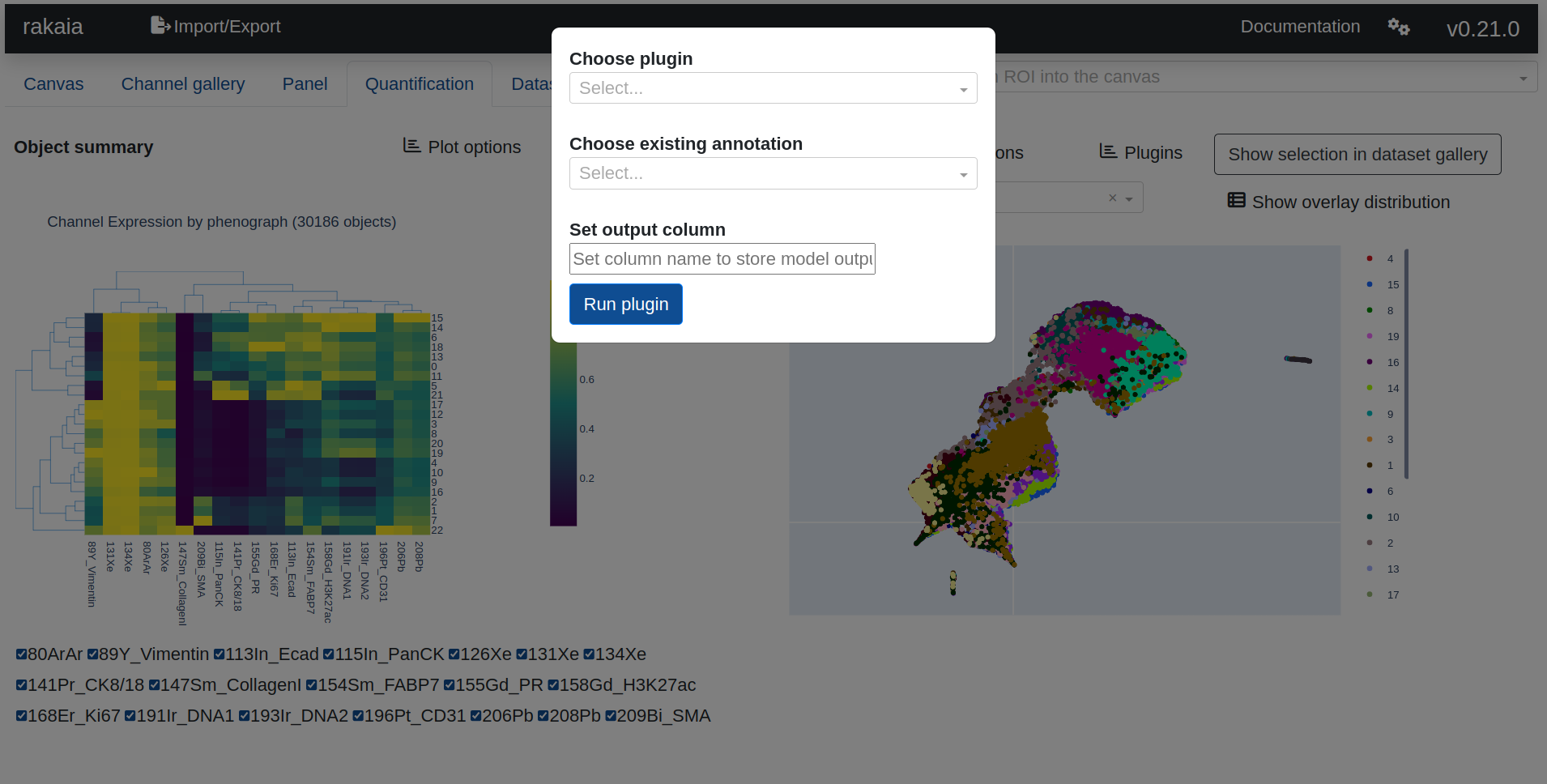
Every model requires the user to specify an output "column" that will store the outputs of the model, which typically includes a label classification oer pbject to be processed. Certain models will also require an input column, which will take the existing labels in the column and use them during training. The following models require an input column:
- Random forest classification for pre-annotated object/structures
Once a model has completed, the new output column will appear as a selection in the UMAP overlay dropdown. This overlay can then be selected as a projection on top of the UMAP and will populate the channel expression summary chart accordingly.
Plugin Support
In development